Protein Purification
Protein purification employs multiple chromatography techniques that separate products according to differences of their properties. Tagged proteins are convenient to be handled by affinity chromatography, which is designed to capture the target protein based on biorecognition of the protein tag. Other protein purification methods, including ion exchange chromatography, size-exclusion chromatography, and hydrophobic interaction chromatography are available if you are interest in obtaining tag-free proteins or products with high purity.
With state-of-art technical platforms for recombinant protein purification, high quality products tailored to your needs are delivered in a short time frame. We are delight to work with you if you have special requirements or protocols for your projects.
Selection guide of protein purification methods
| Protein purification techniques |  |
 |
 |
 |
|---|---|---|---|---|
| name | Affinity chromatography | Ion exchange chromatography | Size-exclusion chromatography | Hydrophobic interaction chromatography |
| Protein property | Biorecognition | Charge | Size | Hydrophobicity |
| Applications | Receptor and ligand, enzyme and substrate, antigen and antibody | Charged molecules | Large molecules, macromolecular complexes | Proteins and peptides with hydrophobic amino acid side chains on surfaces |
| Advantages |
|
|
|
|
| Disadvantages | Demand ligand with high selectivity | Inconsistency from column to column | Demand differences in MW | Too strong interactions |
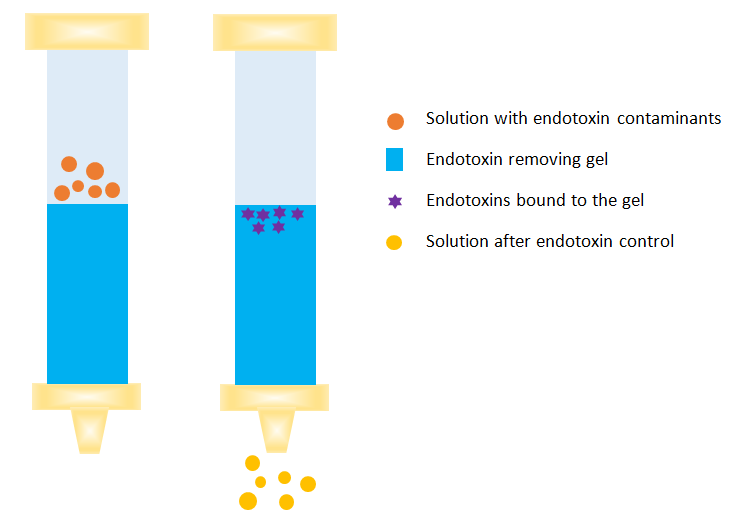
Endotoxin removal in protein purification process
Endotoxin removal is one of the most challenging tasks in the process of protein purification. Multiple techniques are frequently employed to remove endotoxins from protein solutions regard to specific properties of endotoxins in aqueous solution.
| Methods | Key features |
|---|---|
| Affinity chromatography | High selectivity, highly specific and efficient endotoxin removal combined with excellent target protein recovery |
| Ion exchange chromatography | Rapid separation, wide selection of AEC media, sodium hydroxide (NaOH) sanitization, and solvents-free |
| Hydrophobic chromatography | Interacts with non-polar protein surfaces by van der Waals forces |
| Size-exclusive chromatography | Uses highly porous composite polyacrylamide as the column |
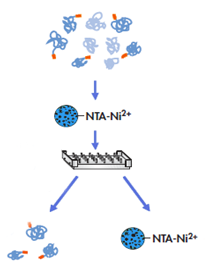
One-step and automated protein purification
Magnetic agarose beads are considered as one of the optimized strategies for one-step and automated protein purification process. Target protein irreversibly binds to the beads and enables efficient protein capture. The strategy eliminates protein loss in of wash steps and results in high protein recovery.
Combination of protein purification strategies
Single step protein purification is frequently insufficient to achieve desired level of purity. The choices of protein purification methods depend greatly upon products properties. Combinations of several classical protein purification techniques are also employed to achieve request purity of proteins without affinity tags.
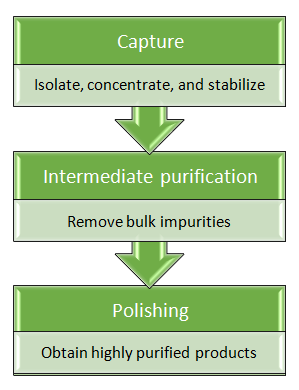
| Protein purification methods | Capture | Intermediate | Polish |
|---|---|---|---|
| Affinity chromatography | +++ | +++ | ++ |
| Ion exchange chromatography | +++ | +++ | +++ |
| Size-exclusion chromatography | + | + | +++ |
| Hydrophobic Interaction Chromatography | ++ | +++ | + |
Notes:
- Combine protein purification techniques that apply very different separation mechanisms.
- Keep balance between purity and yield
Trouble shooting guide of protein purification
| Problems | Possible causes | Recommendation |
|---|---|---|
| Target protein not bound to the column | Protein degradation or tag loss |
|
| Tag inaccessible (unexposed) |
|
|
| Unoptimized binding conditions |
|
|
| Poor target protein elution | Too mild elution conditions |
|
| Too strong interaction with the column |
|
|
| Precipitated target protein | Add solubilizing agents | |
| Presence of non-specific hydrophobic or other interactions | Change imidazole concentration in gradient elution | |
| High amount of co-eluted contaminants | Unoptimized binding conditions |
|
| High concentrated or viscosity sample |
|
|
| Insufficient wash |
|
|
| Multiple bands observed after WB | Partial degradation | Add protease inhibitors |
| Associated contaminants |
|
|
| Cell disruption |
|
|
| Gray or brown resin | Presence of reducing agents |
|
| Column clogged | Presence of cell debris | Centrifuge or filter the sample |
| Protein precipitation |
|
|
| Inclusion body formation | Please see flexible refolding strategies |
The commonly used FDA-approved techniques for endotoxin detection are the rabbit pyrogen test and Limulus Amoebocyte Lysate (LAL) assay.
| Tag | Size | Matrix | Elution condition |
|---|---|---|---|
| His-tag | 6-10 His | Ni2+-NTA, Co2+-CMA | Imidazole 20–250 mM or low pH |
| Poly-Arg | 5-6 Arg | Cation-exchange resin | NaCl linear gradient from 0 to 400 mM at alkaline pH>8.0 |
| GST (Glutathione-S-transferase) | 211 aa | Glutathione | 5–10 mM reduced glutathione |
| MBP (maltose-binding protein) | 396 aa | Cross-linked amylose | 10 mM maltose |
| S-tag | 15 aa | S-fragment of RNaseA | 3 M guanidine thiocyanate |
| FLAG tag | 8 aa | Anti-FLAG monoclonal antibody | pH 3.0 or 2–5 mM EDTA |
| Strep-tag | 8 aa | Strep-Tactin (modified streptavidin) | 2.5 mM desthiobiotin |
| c-myc-tag | 11 aa | Monoclonal antibody | Low pH |
| Cellulose-binding domain | 27–189 aa | Cellulose | Family I: guanidine HCl or urea>4 M Family II/III: ethylene glycol |
| Calmodulin-binding peptide | 26 aa | Calmodulin | EGTA or EGTA with 1 M NaCl |
Case study of flexible protein purification strategies
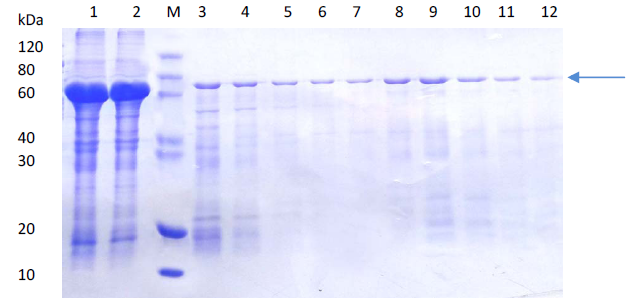 (a)
(a)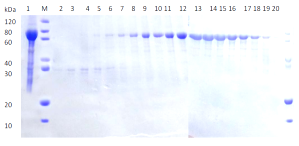 (b)
(b)Comparison of SDS-PAGE analysis of target protein purification
(a) Protein purification using affinity chromatography
Lane M: Protein marker
Lane 1: Supernatant after centrifugation
Lane 2: Flow through
Lane 3-7: 50 mM imidazole eluted fractions
Lane 8-10: 100 nM imidazole eluted fractions
Lane 11-12: 300 nM imidazole eluted fractions
(b) Protein purification using size exclusion chromatography
Lane M: Protein marker
Lane 1: Supernatant after centrifugation
Lane 2-20: Flow through
Contact Us